The High Value Chemicals in Plants (HVCfP) Network in a BBSRC-funded NIBB (Network in Industrial Biotechnology and Bioenergy) that is centered at the University of York. At the end of October, Cambridge Cottage at Kew Gardens hosted the HVCfP Annual Meeting that brought together over 80 interested members of the network to discuss recent research developments and their resulting commercial potential.
The schedule was comprised of invited keynote speakers as well as talks from contributors who have taken advantage of the funding opportunities provided by the network. These include the Proof of Concept (PoC) fund that are worth up to £50K as well as smaller Business Innovative Vouchers (BIV) (previously £5K, now increased to £10K). Funding opportunities are ongoing through the life of the NIBB so please check out funding (https://hvcfp.net/funding/) page for information about previous awards and current open calls.
The meeting kicked off with two keynote talks, firstly from Monique Simmons, deputy director of Kew Science who introduced the wide variety of resources associated with the Botanic Gardens. Over the past year, Kew has undergone a significant reorganization and has a renewed remit for interaction with the scientific community. Perhaps of greatest interest to GARNet readers will be the Kew DNA bank (http://apps.kew.org/dnabank/homepage.html) that contains over 40K samples from around the globe, the majority of which are available to order, either in aliquots suitable for PCR or in larger quantities if necessary.
David Hughes from Syngenta followed up with an excellent talk about the current situation surrounding the debate around Genetically Modified Crops. He gave his thoughts on why this ‘debate’ has gone on for almost 20 years even without any reliable evidence about the negative effects of developed crops. He thought the negative public response could be summed up by three emotions.
Essentialism: is the ‘essence’ of the original organism transferred to the crop
Naturalism: ‘Synthetic’ is evil, ‘natural’ is good.
Disgust: If something is not ‘100% safe’, then it must be bad.
David also advised that the power of narrative is a much more effective technique than the use of potentially bland scientific evidence, which usually falls on deaf ears amongst the public.
The day included two talks from commercial members of the HVCfP network that provided a contrast to the majority academic talks. They included a presentation from James King of Oxford Biotrans (http://oxfordbiotrans.com/), a company that is attempting to exploit biocatalytic routes to develop novel compounds. Their focus is on the use of cytochrome P450s and one of their initial challenges is to enzymatically produce nootkatone, which provides the flavor and scent from grapefruit. They aim to produce this compound via the intermediate Valecene, which can be collected from oranges. The unpredictable grapefruit harvest and the difficulties in isolation of endogeous nookatone mean that Oxford BioTrans suggest that using enzyme catalysis might be a more cost effective method for isolation of the high value product. The other SME that presented was Neem Biotech, which is located in the South Wales Valleys, a place not usually associated with biotechnology. Their major product is Vetrinol, which includes garlic extract and that is used for boosting the bovine immune system.
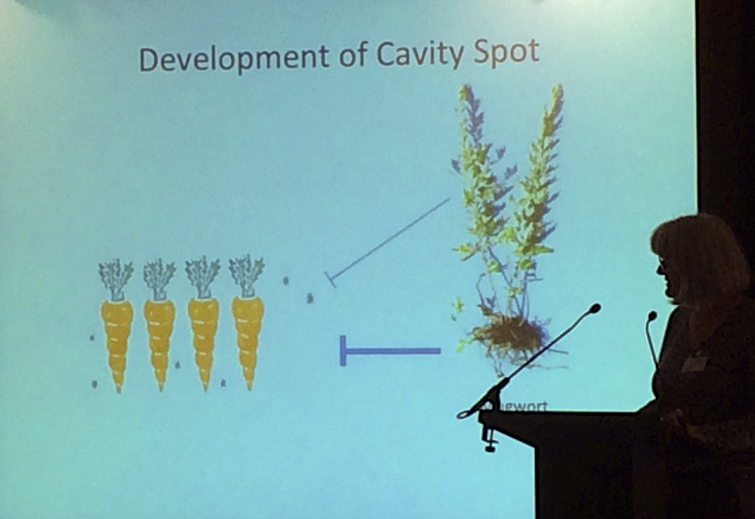
The remainder of talks featured researchers involved in PoC or BIV projects. These were all rather fascinating (especially for a researcher usually preoccupied by Arabidopsis) as they introduced novel compounds and non-model organisms that were ‘unusual’ to many in the audience. Cathie Martin (JIC) is well known for her work with purple tomatoes but here she presented her attempts to isolate the compound from Mugwort leaf extract that is responsible for reducing cavity root spot disease, which is caused by Pithium species of Oomycete. This work was performed in collaboration with ‘Root Crop Consultancy’ and resulted from an observation that carrots grown in the presence of mugwort were less susceptible to cavity spot disease.
Three talks that described research funded by PoC grants came from Gary Loake (Edinburgh), Luis Mur (Aberystwyth) and Mike Beale (Rothamstead) who each highlighted work aimed at to improve production from organisms already used to produce medicinally useful compounds. Gary Loake is hoping to develop the use of cambial meristematic cells (CMC) to improve paclitaxel biosynthesis from Taxus (Yew) (for review of CMCs http://www.sciencedirect.com/science/article/pii/S1871678415000205). The Taxus CMCs are able to produce plenty of paclitaxel so to improve this aspect they have analysed expression profiles in an attempt to identify the transcription factors that are important in this process. The ultimate aim would be to use synthetic biology approaches to potentially modify the expression of these TFs in order to maximise paclitaxel production.
Aberystwyth University has a strong tradition of research in Avena species (Oat) so Dr Mur is using these resources in order to understand and maximise the production of Avenanthramide, which has well-established medicinal benefits. The HVCfP money has funded the use of HPLC to analyse their germplasm for altered production of Avenanthramide. Broadly they have found that domesticated hexaploid Avena species have higher levels of the compound compared to wild diploid varieties. They are now conducting expression analysis in order to understand the molecular reason for these alterations.
Mike Beale is taking advantage of the National Willow Collection (NWC) to undertake a phytochemical and pharmacological screen in order to identify novel compounds. Willow (Salix) is famously the original source of aspirin so Dr Beale and colleagues are using NMR and MS in an attempt to analyse unpurified extracts from 200 species of Willow. By screening against a battery of cancer cell-lines, fungi and bacteria, they hope to identify extracts that can be further interrogated to find compounds of interest.
The other two talks resulting from BIV-funding were both focused on algae, although with organisms of different sizes! David Bailey from IOTA Pharmaceuticals outlined their plans to exploit the potential of using the macroalgae Sargassum muticum (seaweed) as a production chassis while Paul Knox (Leeds) and John Dodd (AlgaeCytes) described their early work to identify novel polysaccharides found in microalgae.
The meeting ended with a series of flash presentations given by representatives of GARNet, the Liverpool GeneMill, the University of Essex Cell screening facility and finally from Matt Davey (Cambridge). His talk highlighted the incredible potential that is provided by plant species as he is developing commercial relationships to study the UV absorption properties of Living Stone species (Lithops). Initial experiments have shown that direct UV light is very successfully absorbed by Lithops species and this is of interest to the cosmetic industry.
Although much of the research presented in this meeting is in its infancy, the HVCfP Network can be pleased with the process it has made. With another set of PoC and BIV funding recently announced the network is supporting research in a wide range of organisms aimed at producing a varying array of compounds. The network has been providing funding since 2014 so the first set of PoC funding is now coming to an end. The true success of the network will be realised over the next two years, determined by the amount of follow-on funding that can be obtaining to support the research conducted in these seed-projects. We will watch this space with interest….