This weeks Arabidopsis Research Roundup begins with two papers from Royal Hollaway University of London that investigate the factors that control leaf development in the dark and the control of PIN1 phosphorylation. Third is a paper from Bristol that demonstrates the translation of research from Arabidopsis into coriander with regard the control of the response to UV light. Next is research from the John Innes Centre that characterises the role of DNA methylation during meiosis in the male lineage.
Christine Foyer (Leeds) leads the next paper that defines the relationship between cold treatment and strigolactone signalling. The penultimate paper is led by Richard Napier from the University of Warwick and determines the parameters that define the substrates of the AUX1 protein whilst the final paper includes Cyril Zipfel (TSL) as a co-author and uses systems biology approaches to characterise the interactions between leucine-rich repeat receptor kinases (LRR-RKs).
Mohammed B, Farahi Bilooei S, Doczi R, Grove E, Railo S, Palme K, Ditengou FA, Bögre L, Lopez-Juez E (2017) Converging energy and hormonal signalling control meristem activity, leaf initiation and growth Plant Phys doi: 10.1104/pp.17.01730
Open Access
Enrique Lopez-Juez (RHUL) leads this collaboration with German and Hungarian colleagues that investigates the fundamental question; ‘Why don’t leaves grow in the dark’. They show that this response is influenced by both auxin transport and the plants energy sensing mechanisms. Interestingly when energy is provided via external sucrose, leaves develop differently in the dark than they do in the light indicating that multiple signaling pathways differentially influence this phenotype.
Enrique discusses this paper on the GARNet YouTube page.
Dory M, Hatzimasoura E, Kállai BM, Nagy SK, Jäger K, Darula Z, Nádai TV, Mészáros T, López-Juez E, Barnabás B, Palme K,,, Bögre L, Ditengou FA,,, Dóczi R (2017) Coevolving MAPK and PID phosphosites indicate an ancient environmental control of PIN auxin transporters in land plants. FEBS Lett. doi: 10.1002/1873-3468.12929
Laszlo Bogre and Enrique Lopez-Juez (RHUL) are co-authors on this Hungarian-led study that has discovered 3 conserved putative MAPK sites within the auxin transport protein PIN1. Phosphorylation of two of these sites causes partial loss of PIN1 membrane localization and therefore opposes the effect of the PINOID kinase, whose activity promotes PIN1 membrane localisation.
Fraser DP, Sharma A, Fletcher T, Budge S, Moncrieff C, Dodd AN, Franklin KA (2017) UV-B antagonises shade avoidance and increases levels of the flavonoid quercetin in coriander (Coriandrum sativum). Sci Rep. doi: 10.1038/s41598-017-18073-8 Open Access
Keara Franklin and Anthony Dodd (University of Bristol) lead this collaboration between academic researchers and those in the company Vitacress. They translate research from Arabidopsis into Coriander that looks at the effect of UV-B on stem elongation and the interaction with flavonoid signaling. This work shows that alterations to the UV-B regime during growth of potted herbs might reduce deleterious effects caused by neighbour proximity.
Walker J, Gao H, Zhang J, Aldridge B, Vickers M, Higgins JD, Feng X (2017) Sexual-lineage-specific DNA methylation regulates meiosis in Arabidopsis. Nat Genet. doi: 10.1038/s41588-017-0008-5
Xiaoqi Feng (JIC) is the corresponding author on this collaboration with James Higgins from Leicester and they investigate the role of DNA methylation in the control of male meiosis. They demonstrate that RNA-directed DNA methylation (RdDM) in the male lineage regulates gene expression in meiocytes and results in the mis-splicing of the MPS1/PRD2 transcipt, which causes aberrant alterations in spindle formation.
Cooper JW, Hu Y, Beyyoudh L, Yildiz Dasgan H, Kunert K, Beveridge CA, Foyer CH (2018) Strigolactones positively regulate chilling tolerance in pea and in Arabidopsis. Plant Cell Environ. doi: 10.1111/pce.13147
Christine Foyer (Leeds) is the corresponding author on this collaboration with Australian, Turkish and South African colleagues that looks into the role strigolactones play during chilling tolerance in both Arabidopsis and pea plants. Plants that have been chilled during the night have reduced biomass, which was not observed in either pea or Arabidopsis strigolactone mutants. This demonstrates a clear role for this hormone in this response and provides a potential target for the manipulation of plant growth under environmental conditions.
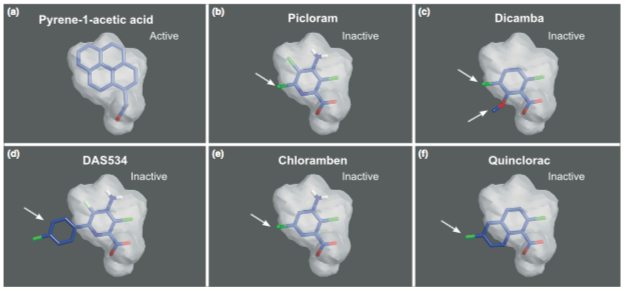
Hoyerova K, Hosek P, Quareshy M, Li J, Klima P, Kubes M,, Yemm AA, Neve P, Tripathi A, Bennett MJ, Napier RM (2017) Auxin molecular field maps define AUX1 selectivity: many auxin herbicides are not substrates. New Phytol. doi: 10.1111/nph.14950
Together with Czech co-authors Richard Napier (Warwick University) leads this investigation into the mode of action of the AUX1 auxin influx carrier and its substrate preferences. This work made use of a novel auxin accumulation assay and associated mathematical modeling to describe the parameters that make difference auxins to be good candidates for the AUX1 transport. Interesting they find that many commonly used auxinicide herbicides are poor substrates for AUX1 and the relevance of this finding for herbicide management strategies.
Smakowska-Luzan E et al (2018) An extracellular network of Arabidopsis leucine-rich repeat receptor kinases. Nature doi: 10.1038/nature25184
Cyril Zipfel (TSL) is a co-author on this US-European study that performs a systems-biology analysis on the possible interactions between extracellular domains of the leucine-rich repeat receptor kinases (LRR-RKs) gene family in Arabidopsis. Analysis of 40K potential interactions allows the generation of a LRR-based cell surface interaction network (CSI-LRR). This was used to discover previously uncharacterized interactions between LRR-RKs and to demonstrate that these interactions allow the translocation of extracellular signals in balanced and tightly regulated patterns.