This weeks UK Arabidopsis Research Roundup features work from two members of the GARNet advisory board who are working on very different aspects of how plants response to external stimuli. In addition there is a genetic and biochemical dissection of primary cell wall formation as well as a comment piece that questions recent findings concerning the relationship between auxin, ABP1 and cortical microtubules.
Busoms S, Teres J, Huang X, Bomblies K, Danku J, Douglas A, Weigel D, Poschenrieder C, Salt DE (2015) Salinity is an agent of divergent selection driving local adaptation of Arabidopsis thaliana to coastal habitats Plant Physiology http://dx.doi.org/pp.00427.2015
Current GARNet Chairman David Salt from Aberdeen has collaborated with researchers from Spain, Germany and the USA in this study that looks at the drivers of adaptive evolution of Arabidopsis plants grown in saline conditions. Unusually this is a field-based study using Arabidopsis that naturally grow in coastal or inland areas of NE Span. Plants taken from coastal areas outperform inland plants when grown on highly saline soils, indicating local adaptation to salt tolerance. The authors conclude that the variation in sodium concentration is causing divergent selection between these two populations.
Monaghan J, Matschi S, Romeis T, Zipfel C (2015) The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst Plant Signaling and Behaviour June 2015 http://dx.doi.org/10.1080/15592324.2015.1018497
GARNet advisory board member Cyril Zipfel from the Sainsbury lab led this study looking at the role of the cytoplasmic kinase BIK1 in the plants response to microbial infection. In plants that are mutant for the Ca2+-dependent protein kinase CPK28, BIK1 accumulates, which leads to enhancing immune signaling. In this study the authors add to these previous finding from their lab by showing that CPK28 also contributes to a burst of Ca2+ production following exposure to pathogens.
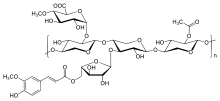
Mortimer JC, Faria-Blanc N, Yu X, Tryfona T, Sorieul M, Ng YZ, Zhang Z, Stott K, Anders N, Dupree P (2015) An unusual xylan in Arabidopsis primary cell walls is synthesised by GUX3, IRX9L, IRX10L and IRX14 Plant Journal http://dx.doi.org/10.1111/tpj.12898
Paul Dupree from the Biochemistry department at the University of Cambridge led this work that investigated a newly characterised form of Xylan, a little studied component of the plant primary cell wall. Genetic analysis indicates that the IRX9L, IRX10L and IRX14 proteins are necessary for xylan backbone synthesis. Importantly this new xylan is contains GlcA side chains, whose addition only requires the glucuronyltransferase GUX3. This type of xylan has not been observed in secondary cell walls so the authors comment on how differences in xylan structure assist in the formation of primary vs secondary cell walls.
T Baskin (2015) Auxin inhibits expansion rate independently of cortical microtubules. Trends in Plant Science http://dx.doi.org/10.1016/j.tplants.2015.05.008
Visiting scholar at CPIB in Nottingham, Tobias Baskin provides a short reply to a publication in Nature that claimed that the control of cell expansion by auxin is caused by reorientation of cortical microtubules. In this paper, Tobias provides evidence from both a simple experiment and from the literature that this might not be the paradigm-shifting observation that it initially appears.