GARNet and OpenPlant were delighted to invite over 140 attendees to the CRISPR-Cas workshop held at the John Innes Centre at the start of September, which was kindly sponsored by Plant Methods. This workshop was designed to introduce CRISPR technology and highlight what might be possible with his transformative technology.
Unfortunately there was a minor hiccup at the start of the day with some confusion over starting time! There was an eager group of researchers beating down the doors at 9am when registration didn’t officially begin until 10am! Hopefully the hour of lost sleep didn’t negatively affect concentration! It was very gratifying to welcome attendees from as far-a-field as Denmark, Ireland, Poland, Germany and Finland! Clearly there is a great appetite to learn about the technology, a fact that might well motivate GARNet to organise another workshop in future, watch this space!
The workshop kicked off with an introduction to OpenPlant from Professor Jim Haseloff before co-organiser Dr Nicola Patron took the attendees through a brief history of gene editing, from the discovery of restriction enzymes in 1975 through to some unpublished gene-editing results from a collaboration with the grass transformation facility at the JIC.
As the focus of the workshop was CRISPR-Cas mediated gene editing it worth reminding readers that this phenomenon was discovered in bacteria as an ‘immune’ response to viral infection. In some undoubtedly ‘Noble-prize’ worthy work it was realised that this could be repurposed for use in eukaryotes as a simple yet precise method of gene editing. In short this takes advantage of the activity of a Cas9 nuclease that is guided to a specific sequence by a 20mer complementary ‘guide-RNA’. Therefore in theory the guide RNA can be designed to target Cas9 to any sequence of interest. This results in the host DNA repair enzymes attempting to fix the error……however this process is not perfect so in many cases a mutation remains, which can abolish or alter gene/protein function.
The first keynote talk was given by Professor Holger Puchta (KIT) who described his seminal work on the use of double strand break (DSB) repair and how this can be combined with gene-editing technologies in plants. Holger described the two most  important molecular discoveries of his lifetime, namely the polymerase chain reaction and gene editing technology, which for the latter he said ‘hit him like a tsunami’ when he realised its enormous potential!
important molecular discoveries of his lifetime, namely the polymerase chain reaction and gene editing technology, which for the latter he said ‘hit him like a tsunami’ when he realised its enormous potential!
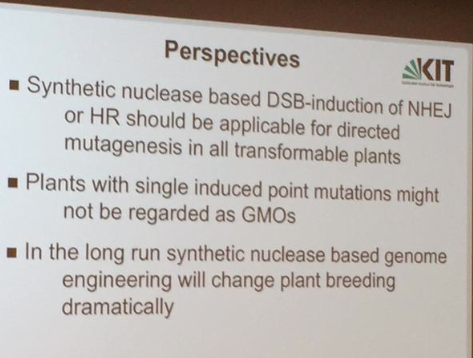
Holger gave an inspiring talk that provided attendees with the history of his work on DSB through to present day applications using CRISPR to induce DSB and homologous recombination for targeted replacement of gene-edited sequences. He ended his talk with some exciting perspectives where he believes that gene-editing technology will dramatically change the future of plant breeding.
Perspective Photo- @BioMatty
After an excellent JIC lunch, the second keynote was provided by Dr Bing Yang from Iowa State where he discussed his work using CRISPR for gene-editing in rice and maize. Bing showed that CRISPR-Cas can be successfully used with high efficiency in monocots which, given the time-consuming transformation procedures in these plants, makes it a very attractive technique for altering gene function. Importantly Bing showed that it was possible to induce bi-allelic mutations in T0 transgenic rice, thus greatly reducing the time to develop plants with altered gene function. 
Arguably the most exciting part of his talk was his final slide in which Bing presented a letter from the US Department of Agriculture (USDA) that stated that his gene-edited rice lines were not considered genetically engineered (and therefore not subject to the usual regulations). If this policy is replicated for other gene edited lines and in other countries then this could be a game-changing development for our ability to use of targeted lines in many areas of plant biology and agriculture…..
The debate surrounding gene-editing will run for a while but the early signs are positive that it will more ‘acceptable’ in the court of public opinion than its related predecessor ‘genetic-modification’.
The later talks were focused on more direct uses of the technology presented by researchers at the Sainsbury Lab, JIC and University of Cambridge. Laurence Tomlinson from Jonathan Jones’s lab presented work where she used CRISPR-Cas to alter the GA signaling pathway to produce dominant dwarf tomatoes. She highlighted that it is impossible to predict the exact nature of the genetic lesion that will occur following CRISPR-Cas targeting, showing tomato plants that were either taller or shorter than wildtype plants due to either loss of function or dominant-negative versions of the protein that they targeted independently by gene editing.
Arguably the clearest talk of the day was given by Vladimir Nekrasov from Sophien Kamoun’s lab who presented a step-wise description of how he used CRISPR-Cas to produce a disease-resistant tomato. He has now selected the CRISPR-Cas transgene out of these edited plants so in theory they are now no more transgenic that any tomato with a random mutation in the gene they targeted. They are hoping to supply these tomato seeds to regions of the world where powdery mildrew is a particular problem and where they will be able to accept the seeds without the bureaucratic hurdles that might exist elsewhere.
The meeting ended with not a little corporate pizzazz from Edward Perello who is a SynBio LEAP fellow and founding member of Desktop Genetics, a company that provides a service to design bespoke CRISPR targets for a users gene of interest. Edward took the opportunity of the meeting to announce that Desktop is now supporting six new plan t genomes with their target design software. He is eager to collaborate with the plant community and encouraged everyone to use the software to design targets and then provide feedback as to their success rate. Importantly this is free to use for academics so if you are looking for software to use as a comparison to your own target design or as a stand alone tool for reliable target prediction then please take the website for a spin!
t genomes with their target design software. He is eager to collaborate with the plant community and encouraged everyone to use the software to design targets and then provide feedback as to their success rate. Importantly this is free to use for academics so if you are looking for software to use as a comparison to your own target design or as a stand alone tool for reliable target prediction then please take the website for a spin!
Overall it was a great day of talks and although there were a few minor issues that came up during feedback (thanks to the third of attendees who provided information), the majority of people found the workshop very useful. GARNet will be providing a PDF of at least some of the talks in the coming weeks so watch this space, the ArabUK newsgroup or the GARNet Facebook page for coming information!