Paper Review: Quantitative characterization of genetic parts and circuits for plant synthetic biology. Nature Methods (2015) doi:10.1038/nmeth.3659
Professor June Medford andd Dr Ashok Prasad (Colorado State) have been in the vanguard of the development of tools for plant synthetic biology over the past few years. Here we highlight a recently published article in Nature Methods that presents their quantification of genetic parts and circuits designed for plant synthetic biology.
To date, quantitatively defined gene circuits have been almost exclusively characterised in unicellular organisms. Some of the initial challenges in transferring this characterisation to multi-cellular organisms includes use of orthogonally useful parts, that are active irrespective of developmental or organismal context, as well as developing methods for the quantification of input and output characteristics. This latter aim is hindered by the difficulties of plant transformation that adds significant levels of variation to this process. In this study the authors aimed to overcome some of these challenges by characterising parts using a medium-throughout method that analyses luciferase expression in protoplasts all within in a 96-well plant format.
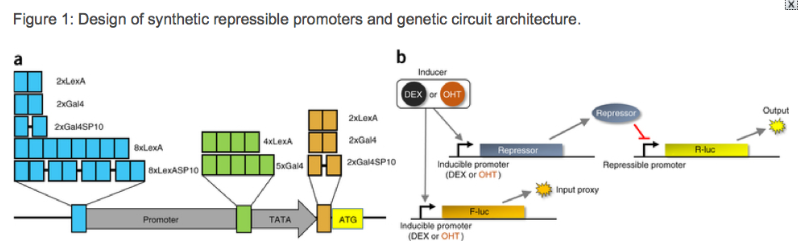
This set of synthetic parts focused on the analysis of different repressive elements fused to a constitutive promotor region. These repressors were designed with orthogonal activity in mind and included both previously characterised and novel elements (Figure 1a). The output utilised a dual luciferase assay where one species of the Luc enzyme was constitutively expressed and compared to the expression level that was influenced by the designed repressor region (Figure 1b). This allowed tuneable expression that is dependent on the type of repressor elements that were present, all within the context of high levels of initial constitutive expression. In total, 128 different promotor combinations were tested in an Arabidopsis protoplast expression system. These expression levels were further varied by altering the levels of induction provided by two different compounds (DEX and OHT).
Initial experimentation generated significant ‘noise’, which is not ideal for generating predictable tools for use in synthetic biology. By analyzing a number of possible sources of experimental variation they found that the ‘batch effect’ was most significant, which described the differences between protoplast samples prepared on different days (attempts to resolve the precise aspect of the experimental technique that caused the variation were unsuccessful). Although it is somewhat harkening to know that the transformation efficiency or uptake of chemical inducers does not contribute as much to experimental variation, it is somewhat worrying that the fundamental preparation of protoplasts provided the highest amount of variation. However the authors were able to develop a mathematical model tat they used in their analysis that accounted for the ‘batch effect’. Ultimately the authors found that the repressor elements worked as expected but that some molecular motives acted more predictably in different pairings, the details of which can be found in the paper.
The authors then attempted to characterise a similar set of constructs in protoplasts generated from the monocot Sorghum. Although a slight alteration was made to the 5’ UTR region of the construct, they found that using the same normalization model to remove the batch effect generated a similar set of results as in the Arabidopsis protoplasts. This indicates that Arabidopsis can act as a good model for evaluating initial expression levels that might occur in other dicots or monocots.
Finally three different promotor-repressor genetic circuits were stably transformed into Arabidopsis and then multiple independent transgenic lines were isolated for each combination. Subsequently one transgenic line from each of three genetic combinations was chosen for comparison with the transient expression system. In a method outlined in Figure 2, protoplasts from the stably transformed plants were compared with those isolated from wildtype pla nts that were transiently transformed with the same construct. Although absolute expression was higher in the transiently transformed protoplasts, analysis of normalized data demonstrated that equivalent repressor elements performed similarly in the differently generated samples. This allowed the authors to suggest that use of the transient system was a reasonable proxy for that observed in stably transformed plants. Therefore they conclude that the transient system can be used as a more rapid screening procedure for newly developed synthetic elements.
nts that were transiently transformed with the same construct. Although absolute expression was higher in the transiently transformed protoplasts, analysis of normalized data demonstrated that equivalent repressor elements performed similarly in the differently generated samples. This allowed the authors to suggest that use of the transient system was a reasonable proxy for that observed in stably transformed plants. Therefore they conclude that the transient system can be used as a more rapid screening procedure for newly developed synthetic elements.
This is an important piece of analysis that is absolutely necessary before the full potential of plant synthetic biology can be realised. This type of design, build, test procedure is common-place when using a unicellular chassis but has not been studied in this detail in a plant system.
However the amount of normalization that is required to ensure the data is directly comparable certainly highlights difficulties may lie ahead as researchers work toward the ultimate goal of developing synthetic genetic elements that are usable across many multi-cellular plant chassis. Although the GARNet blog has highlighted some excellent tools that have been recently developed for plant synthetic biology, this study acts as a cautionary tale for those entering this field and are unaware of the unpredictable nature of even the most commonly used experimental techniques.
Overall there is little doubt that this is an exciting time for plant synthetic biology even though the generation of predictable, consistent gene expression across a variety of chassis will remain rather challenging in the immediate future.