This bumper edition of the GARNet Research Roundup begins with three papers that have a focus on the circadian clock. First is from Cambridge and looks at a novel role for TTG1 in control of the clock. The second paper also includes co-authors from Cambridge and looks at the clock Evening Complex. The final clock paper includes co-authors from York and looks at the new roles for EARLY FLOWERING 3 and GIGANTEA.
The next four papers include researchers from the John Innes Centre. Yiling Ding’s lab lead an exciting study into the role of RNA G-quadruplex to define liquid-liquid phase separations. Next David Seung and Alison Baker look at production of amylose starch across Arabidopsis accessions. The third JIC paper is from the Charpentier lab and looks at nuclear calcium signaling in the root. Finally Lars Ostergaard is a co-author on a paper that identifies a novel biostimulant that controls podshatter in Brassica.
The eighth paper is from Glasgow and describes the bioengineering of plants to express a novel antibiotic bacteriocin.
Next are three papers introduce exciting new research tools. 1. Weibei Yang in the Meyerowitz lab introduces a method for co-labeling of RNAs and protein 2. Researchers in Nottingham introduce RootNav2.0 for the automated measurement of root archtiectures 3. The Haydon Lab has developed a GAL4-GFP luciferase system for tissue-specific gene expression analysis.
Two Photosynthesis-based papers come next with firstly an analysis on the link between metabolism and the light response curve (from Manchester) and secondly a look at the role of aquaporins in control of CO2 conductance (Cambridge and Lancaster).
The fourteenth paper is from Durham and characterises an important protein regulator of the autophagy-dependent degradation pathway whilst the fifteenth is from Cambridge and uses cryo-SEM to analyse cell wall structures.
The penultimate paper is from Birmingham and looks at the role of redox signaling in aphid fecundity and the final paper includes co-authors from RHUL and looks at the interaction between the E2FB and RETINOBLASTOMA-RELATED proteins.
Airoldi CA, Hearn TJ, Brockington SF, Webb AAR, Glover BJ (2019) TTG1 proteins regulate circadian activity as well as epidermal cell fate and pigmentation. Nat Plants. doi: 10.1038/s41477-019-0544-3
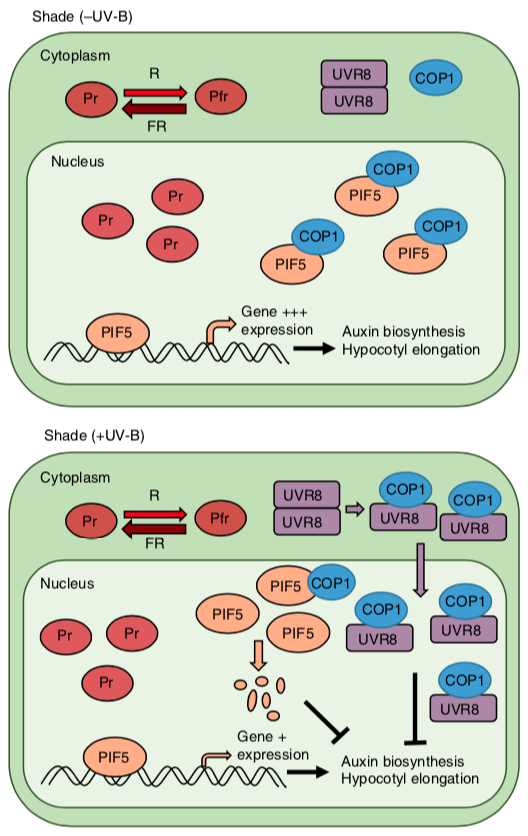
This study from the University of Cambridge is led by Chiara Airoldi and introduces a new role for the TRANSPARENT TESTA GLABRA 1 (TTG1) WD-repeat (WDR) subfamily in the regulation of the circadian clock. TTG1 regulates epidermal cell differentiation and pigment production, while LIGHT-REGULATED WD1 and LIGHT-REGULATED WD2A are known to regulate the clock. The triple lwd1 lwd2 ttg1 mutant has no detectable circadian rhythym. This suggests that members of this protein family have undergone subfunctionalization to diverge from their core functions. This paper is of interest to those who research evolution of protein function as well as the to those interested in the control of the circadian clock.

Tong M, Lee K, Ezer D, Cortijo S, Jung J, Charoensawan V, Box MS, Jaeger K, Takahashi N, Mas P, Wigge PA, Seo PJ (2019) The Evening Complex establishes repressive chromatin domains via H2A.Z deposition. Plant Physiol. doi: 10.1104/pp.19.00881
This collaboration between the UK and South Korea is led by Meixuezi Tong and investigates how the Evening Complex (EC) component of the circadian clock interacts with chromatin to control gene expression at dusk. This occurs through direct interaction with the SWI2/SNF2-RELATED complex and together they bind to the core clock genes PRR7 and PRR9, causing the deposition of H2A.Z at these loci subsequent to causing their repression at dusk.
Anwer MU, Davis A, Davis SJ, Quint M (2019) Photoperiod sensing of the circadian clock is controlled by EARLY FLOWERING 3 and GIGANTEA. Plant J. doi: 10.1111/tpj.14604
Amanda Davies and Seth Davies from the University of York are co-authors on this German-led study with Muhammad Anwer as both first and corresponding author. They look at the role of important circadian regulators ELF3 and GIGANTEA through generation of previously unanalysed elf3gi double mutants. In these plants the circadian oscillator fails to synchronize to light-dark cycles even under diurnal conditions, demonstrating that these genes act together to convey photoperiod sensing to the central oscillator.
Zhang Y, Yang M, Duncan S, Yang X, Abdelhamid MAS, Huang L, Zhang H, Benfey PN, Waller ZAE, Ding Y (2019) G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res. doi: 10.1093/nar/gkz978
Open Access
Yueying Zhang is the first author of this study conducted in the lab of Yiliang Ding at the John Innes Centre, in collaboration with the Benfey lab in the USA. They reveal an exciting mode of regulating RNA activity through the formation of RNA G-quadruplex (GQ) complexes. They use the SHORTROOT mRNA as the model for this study, showing that GQ-mediated complex formation can bring liquid-liquid phase separation. This study is of fundamental importance as it provides the first evidence that RNA can adopt structural motifs to trigger and/or maintain the specificity of RNA-driven phase separation.

Seung D, Echevarría-Poza A, Steuernagel B, Smith AM (2019) Natural polymorphisms in Arabidopsis result in wide variation or loss of the amylose component of starch. Plant Physiol. doi: 10.1104/pp.19.01062
Open Access
David Seung at the John Innes Centre the first and corresponding author of this study that used data from the Arabidopsis 1135 Genome project to investigate the prevelance of amylose production. Plants with amylose-free starch have no detrimental phenotypes so the function of this glucose-polymer, that accounts for up to 30% of all natural starch, is unknown. They looked at the polymorphisms within the GRANULE-BOUND STARCH SYNTHASE (GBSS) enzyme, identifying natural accessions that have no GBSS activity yet are viable within their natural environments. This study is a prelude to future research that will discover the adaptive significance of amylose.
Leitão N, Dangeville P, Carter R, Charpentier M (2019) Nuclear calcium signatures are associated with root development. Nat Commun. doi: 10.1038/s41467-019-12845-8
Open Access
Nuno Leitao is first author on this research from the Charpentier lab at the John Innes Centre. They looked at the role of nuclear Ca2+ signalling on primary root meristem development and auxin homeostasis through activity of the nuclear membrane localised ion channel DOES NOT MAKE INFECTIONS 1 (DMI1). This study discovers a previously unappreciated role for intracellular Ca2+ signalling during plant development.
Łangowski Ł, Goñi O, Quille P, Stephenson P, Carmody N, Feeney E, Barton D, Østergaard L, O’Connell S (2019 A
plant biostimulant from the seaweed Ascophyllum nodosum (Sealicit)
reduces podshatter and yield loss in oilseed rape through modulation of
IND expression. Sci Rep. doi: 10.1038/s41598-019-52958-0
Open Access
Lars Ostergaard is a co-author on this Irish-study led by Lukasz Łangowski that investigates the factors that control pod shatter in oil seed rape. They show that the seaweed Ascophyllum nodosum-based biostimulant (Sealicit) is able to reduce podshatter by effecting the expression of the major regulator of pod shattering, INDEHISCENT. This has implications for the use of this compound by farmers wanting to reduce the amount of seed loss due to premature pod shatter.
Rooney WM, Grinter RW, Correia A, Parkhill J, Walker DC, Milner JJ (2019) Engineering bacteriocin-mediated resistance against the plant pathogen Pseudomonas syringae. Plant Biotechnol J. doi: 10.1111/pbi.13294
Open Access
William Rooney at the University of Glasgow is lead author on this study that attempts to combat Pseudomonas syringae infections through expression of a novel protein antibiotic bacteriocin, putidacin. They show that transgenic expression of this bacterial protein provides effective protection against Pseudomonas. This proof of concept opens the possibility for more widespread use of bacteriocins as an effective plant protection strategy.

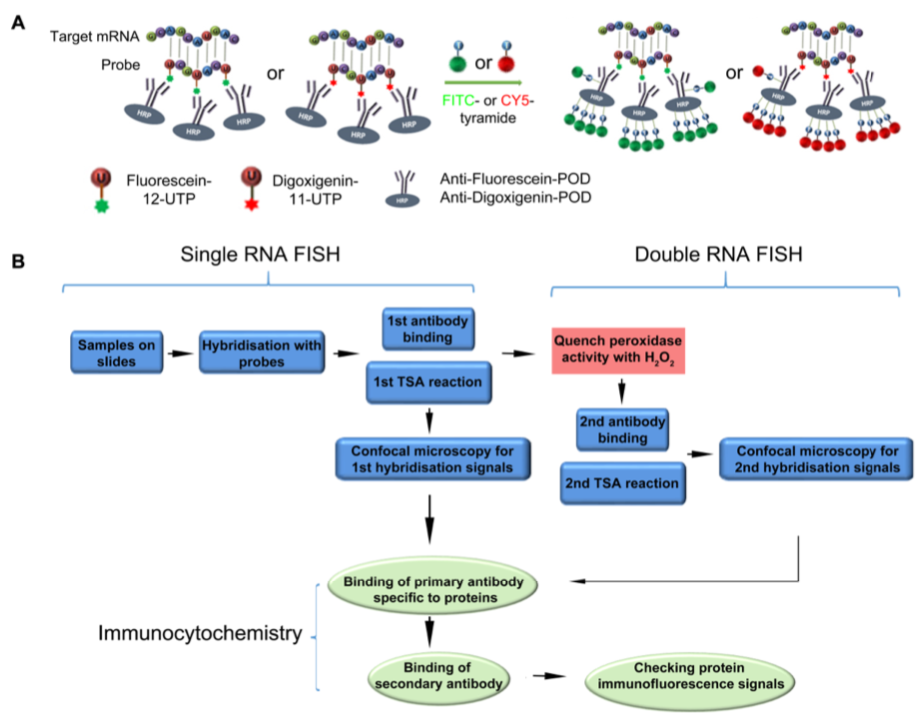
Yang W, Schuster C, Prunet N, Dong Q, Landrein B, Wightman R, Meyerowitz EM (2019) Visualization of Protein Coding, Long Non-coding and Nuclear RNAs by FISH in Sections of Shoot Apical Meristems and Developing Flowers. Plant Physiol. doi: 10.1104/pp.19.00980
This extended methods paper is led by Weibing Yang at the Sainsbury lab in Cambridge. They have adapted RNA fluorescence in situ hybridization (rnaFISH) to explore RNA localization in the shoot apical meristem of Arabidopsis. They are able to label mRNA as well as long ncRNAs and have developed double labeling to assay two separate RNAs in the same cell and to assess nucleo-cytoplasmic separation of RNA species. Finally they link rnaFISH with fluorescence immunocytochemistry for the simultaneous localization of a single genes mRNA and protein.
Yasrab R, Atkinson JA, Wells DM, French AP, Pridmore TP, Pound MP (2019) RootNav 2.0: Deep learning for automatic navigation of complex plant root architectures. Gigascience. doi: 10.1093/gigascience/giz123
Open Access
Robail Yasrab is lead author on this work from the University of Nottingham that introduces the RootNav2.0 software tool. This was developed by modern deep-learning approaches and allows the fully automated measurement of vertically growth root systems. RootNav2.0 was favourably compared with its semi-automated predecessor RootNav1.0 and can be used for measurement of root architectures from a range of different plant species.
Román Á, Golz JF, Webb AA, Graham IA, Haydon MJ (2019) Combining GAL4 GFP enhancer trap with split luciferase to measure spatiotemporal promoter activity in Arabidopsis. Plant J. doi: 10.1111/tpj.14603
This technical advance is led by Angela Roman, was in the Haydon lab during its time at the University of York. They have used the GAL4-GFP enhancer trap system, to develop a tissue-specific split luciferase assay for non-invasive detection of spatiotemporal gene expression in Arabidopsis. In this example they use the study to measure dynamics of circadian gene expression but is clearly applicable to answer many other experimental questions.
Herrmann HA, Schwartz JM, Johnson GN (2019) From empirical to theoretical models of light response curves – linking photosynthetic and metabolic acclimation. Photosynth Res. doi: 10.1007/s11120-019-00681-2
Open Access
Helena Herrmann is lead author on this work fro the University of Manchester. In this study they developed and then empirically tested a series of simple kinetic models that explains the metabolic changes that are required to alter light response curves (LRCs) across a range of temperatures. This allowed them to show how changes in NADPH and CO2 utilization respond to environmental changes. This provides useful information as to how a plant adapts its metabolic response to light depending on the growth temperature.
Kromdijk J, Głowacka K, Long SP (2019) Photosynthetic efficiency and mesophyll conductance are unaffected in Arabidopsis thaliana aquaporin knock-out lines. J Exp Bot. doi: 10.1093/jxb/erz442
Open Access
Wanne Kromdijk
leads this US-led research that includes contributions from the
Universities of Cambridge and Lancaster. They looked at the potential
role of membrane-bound aquaporins in the control of diffusion
conductance for CO2 transfer from substomatal cavity to chloroplast
stroma (gm). They tested three aquaporin mutants across a range of
light and CO2 concentrations and surprisingly found that they appear to
play no significant contribution to the control of gm. The reporting of
this type of ‘negative’ result will prevent unnecessary replication of experiments and help to streamline the research process.
Wang
P, Pleskot R, Zang J, Winkler J, Wang J, Yperman K, Zhang T, Wang K,
Gong J, Guan Y, Richardson C, Duckney P, Vandorpe M, Mylle E, Fiserova
J, Van Damme D, Hussey PJ (2019) Plant AtEH/Pan1 proteins drive autophagosome formation at ER-PM contact sites with actin and endocytic machinery. Nat Commun. doi: 10.1038/s41467-019-12782-6
Open Access
Pengwei Wang is first author in this research led from Durham University that incudes Chinese and Belgian collaborators. They show that the AtEH/Pan1 protein is involved with actin cytoskeleton regulated autophagy and recruits multiple other components to autophagosomes during this process. In addition they show vesicle bound-AtEH/Pan1 interact with VAP27-1 at the ER-PM. This demonstrates that AtEH/Pan1 is a key component of the autophagy-dependent degradation pathway.
Lyczakowski JJ, Bourdon M, Terrett OM, Helariutta Y, Wightman R, Dupree P (2019) Structural Imaging of Native Cryo-Preserved Secondary Cell Walls Reveals the Presence of Macrofibrils and Their Formation Requires Normal Cellulose, Lignin and Xylan Biosynthesis. Front Plant Sci. doi: 10.3389/fpls.2019.01398
Open Access
Jan Lyczakowski from the Dupree lab at the University of Cambridge is first author on this study that has adapted low temperature scanning electron microscopy (cryo-SEM) to visualize the cell walls of both angiosperm and gymnosperms. They have used Arabidopsis mutants to reveal that cell wall macrofibrils at composed of cellulose, xylan, and lignin. They demonstrate that cryo-SEM is a useful tool for native nanoscale cell wall architectures.
Rasool B, Karpinska B, Pascual J, Kangasjärvi S, Foyer CH (2019) Catalase,
glutathione and protein phosphatase 2A-dependent organellar redox
signalling regulate aphid fecundity under moderate and high irradiance. Plant Cell Environ. doi: 10.1111/pce.13669
Brwa
Rasool is first author on this collaboration between the Universities
of Birmingham and Helsinki that looks at how aphids respond to redox
changes in Arabidopsis thaliana grown under different light
conditions. They also identified defence-related transcription factors
differentially upregulated by aphid predation in different light
conditions. Overall they show aphid fecundity is in part determined by
the plants cellular redox signaling.
Őszi E, Papdi C, Mohammed B, Pettkó-Szandtner A, Vaskó-Leviczky T, Molnár E, Ampudia CG, Khan S, Lopez-Juez E, Horváth B, Bögre L, Magyar Z (2019) E2FB interacts with RETINOBLASTOMA RELATED and regulates cell proliferation during leaf development. Plant Physiol. doi: 10.1104/pp.19.00212
Erika Oszi is first author of this Hungarian-led research that includes co-authors from Royal Holloway University of London. This research looks at the interaction between the transcription factors E2FB and RETINOBLASTOMA-RELATED (RBR) and how this contributes to cell proliferation during organ development in Arabidopsis leaves. The relationship between these proteins changes throughout the stages of leaf development and is critical to determine final leaf cell number.